AeMCMC#
AeMCMC is a compiler for Bayesian inference.
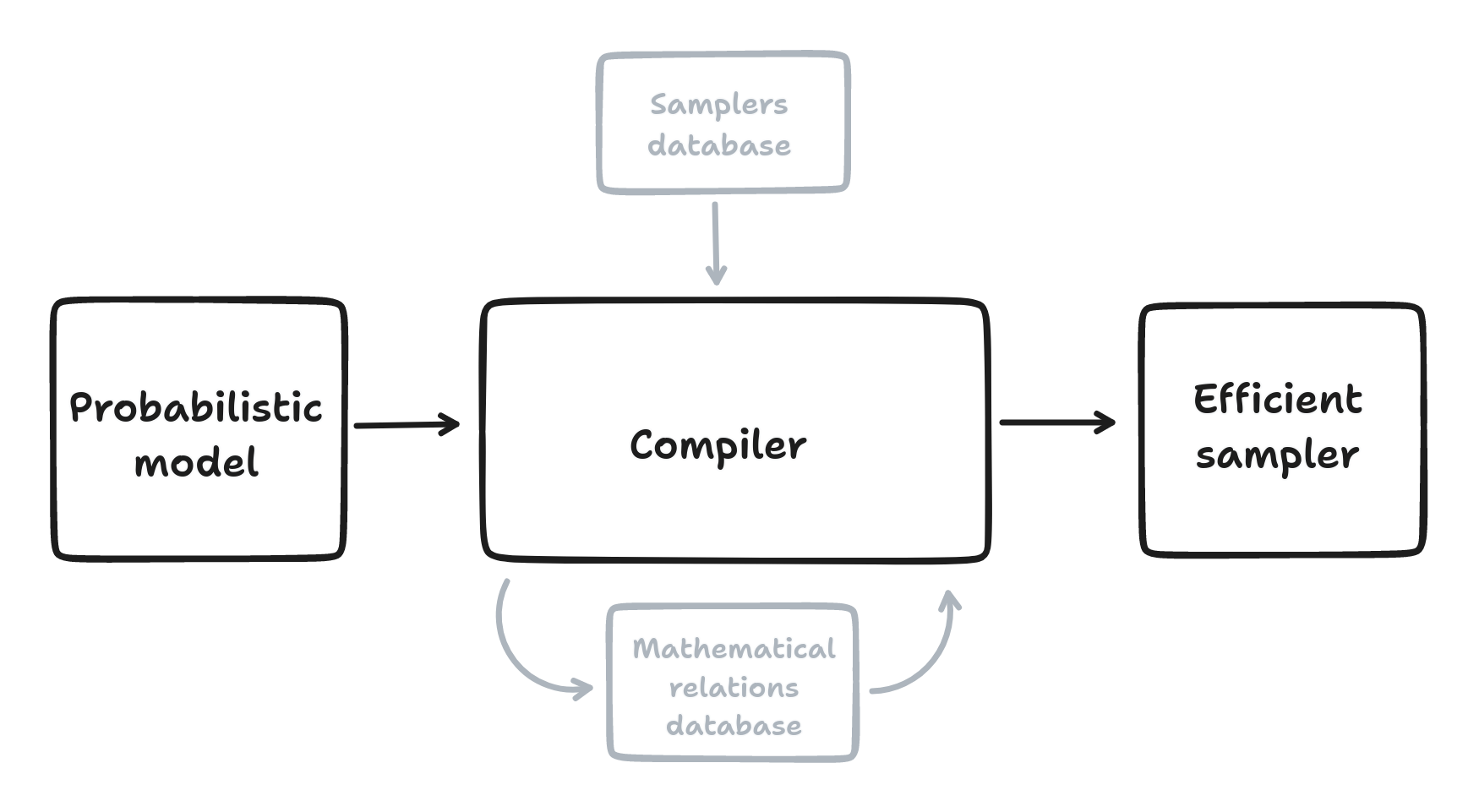
AeMCMC is a Python library that automates the construction of samplers for probabilistic models built with Aesara. AeMCMC analyzes the model graph and possibly rewrites it to find the most suitable samplers.
Features#
Automatically identify and compose samplings steps
Database of mathematical relations
Database of “exact” posterior sampling steps and general-purpose samplers like NUTS
We aim to provide a user-friendly interface similar to that of other PPLs, but with much faster and more efficient inference.
Example#
In this example we show that AeMCMC can recognize closed-form posteriors. For instance, the following Beta-Binomial model amounts to sampling from a Beta distribution:
import aesara
import aemcmc
import aesara.tensor as at
srng = at.random.RandomStream(0)
p_rv = srng.beta(1., 1.)
Y_rv = srng.binomial(10, p_rv)
y_vv = Y_rv.clone() # observations
sampler, initial_values = aemcmc.construct_sampler({Y_rv: y_vv}, srng)
p_posterior_step = sampler.sample_steps[p_rv]
sample_fn = aesara.function([y_vv], p_posterior_step)
AeMCMC has succesfully identified the conjugacy relation, and \(p\) is given by a \(\operatorname{Beta}\) distribution:
aesara.dprint(p_posterior_step)
# beta_rv{0, (0, 0), floatX, False}.1 [id A]
# |RandomGeneratorSharedVariable(<Generator(PCG64) at 0x7F77B2831200>) [id B]
# |TensorConstant{[]} [id C]
# |TensorConstant{11} [id D]
# |Elemwise{add,no_inplace} [id E]
# | |TensorConstant{1.0} [id F]
# | |y [id G]
# |Elemwise{sub,no_inplace} [id H]
# |Elemwise{add,no_inplace} [id I]
# | |TensorConstant{1.0} [id F]
# | |TensorConstant{10} [id J]
# |y [id G]
Install AeMCMC#
AeMCMC installation can happen in a few different ways. You can install AeMCMC with conda or with pip. To get the bleeding edge version you can install aemcmc-nightly.
pip install aemcmc
conda install -c conda-forge aemcmc
pip install aemcmc-nightly
